Validation
A number of functions are made available to assist in validation of the estimated models. We illustrate by an example
Generate some test data:
using ControlSystemIdentification, ControlSystemsBase, Random
using ControlSystemIdentification: newpem
Random.seed!(1)
T = 200
nx = 2
nu = 1
ny = 1
x0 = randn(nx)
σy = 0.5
sim(sys,u) = lsim(sys, u, 1:T)[1]
sys = tf(1, [1, 2*0.1, 0.1])
sysn = tf(σy, [1, 2*0.1, 0.3])
# Training data
u = randn(nu,T)
y = sim(sys, u)
yn = y + sim(sysn, randn(size(u)))
dn = iddata(yn, u, 1)
# Validation data
uv = randn(nu, T)
yv = sim(sys, uv)
ynv = yv + sim(sysn, randn(size(uv)))
dv = iddata(yv, uv, 1)
dnv = iddata(ynv, uv, 1)InputOutput data of length 200, 1 outputs, 1 inputs, Ts = 1We then fit a couple of models
res = [newpem(dn, nx, focus=:prediction) for nx = [2,3,4]];Iter Function value Gradient norm
0 8.240162e+01 4.646640e+02
* time: 4.291534423828125e-5
50 1.383464e+01 6.849784e+01
* time: 0.017041921615600586
Iter Function value Gradient norm
0 6.829949e+01 5.815865e+02
* time: 4.291534423828125e-5
50 1.085330e+01 9.525272e+00
* time: 0.04268693923950195
100 9.120639e+00 2.471301e-02
* time: 0.05151796340942383
Iter Function value Gradient norm
0 3.349007e+01 1.278532e+02
* time: 4.220008850097656e-5
50 7.636427e+00 2.791433e+01
* time: 0.053559064865112305
100 7.636206e+00 2.789863e+01
* time: 0.10906505584716797After fitting the models, we validate the results using the validation data and the functions simplot and predplot (cf. Matlab sys.id's compare):
using Plots
ω = exp10.(range(-2, stop=log10(pi), length=150))
fig = plot(layout=4, size=(1000,600))
for i in eachindex(res)
sysh, x0h, opt = res[i]
simplot!( sysh, dnv, x0h; sp=1, ploty=false)
predplot!(sysh, dnv, x0h; sp=2, ploty=false)
end
plot!(dnv.y' .* [1 1], lab="y", l=(:dash, :black), sp=[1 2])
bodeplot!((getindex.(res,1)), ω, link = :none, balance=false, plotphase=false, subplot=3, title="Process", linewidth=2*[4 3 2 1])
bodeplot!(innovation_form.(getindex.(res,1)), ω, link = :none, balance=false, plotphase=false, subplot=4, linewidth=2*[4 3 2 1])
bodeplot!(sys, ω, link = :none, balance=false, plotphase=false, subplot=3, lab="True", l=(:black, :dash), legend = :bottomleft, title="System model")
bodeplot!(innovation_form(ss(sys),syse=ss(sysn)), ω, link = :none, balance=false, plotphase=false, subplot=4, lab="True", l=(:black, :dash), ylims=(0.1, 100), legend = :bottomleft, title="Noise model")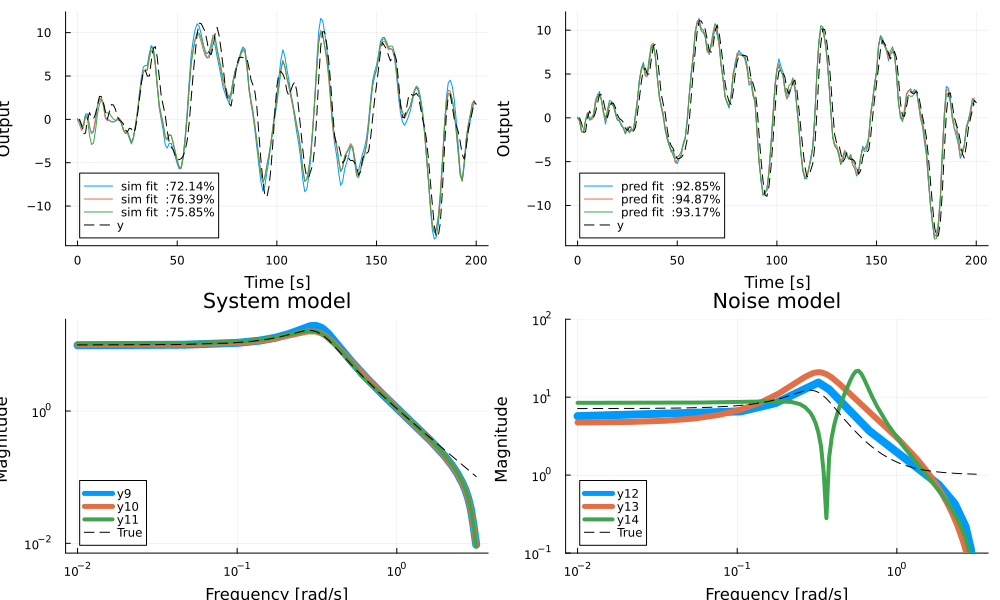
In the figure, simulation output is compared to the true model on the top left and prediction on top right. The system models and noise models are visualized in the bottom plots. All models capture the system dynamics reasonably well, but struggle slightly with capturing the gain of the noise dynamics. The true system has 4 poles (two in the process and two in the noise process) but a simpler model may sometimes work better.
Prediction models may also be evaluated using a h-step prediction, here h is short for "horizon".
figh = plot()
for i in eachindex(res)
sysh, x0h, opt = res[i]
predplot!(sysh, dnv, x0h, ploty=false, h=5)
end
plot!(dnv.y', lab="y", l=(:dash, :black))
fighIt's generally a good idea to validate estimated model with a prediction horizon larger than one, in particular, it may be valuable to verify the performance for a prediction horizon that corresponds roughly to the dominant time constant of the process.
See also simulate, predplot, simplot, coherenceplot
Different length predictors
When the prediction horizon gets longer, the mapping from $u \rightarrow ŷ$ approaches that of the simulation system, while the mapping $y \rightarrow ŷ$ gets smaller and smaller.
using LinearAlgebra
G = c2d(DemoSystems.resonant(), 0.1)
K = kalman(G, I(G.nx), I(G.ny))
sys = add_input(G, K, I(G.ny)) # Form an innovation model with inputs u and e
T = 10000
u = randn(G.nu, T)
e = 0.1randn(G.ny, T)
y = lsim(sys, [u; e]).y
d = iddata(y, u, G.Ts)
Gh,_ = newpem(d, G.nx, zeroD=true)
# Create predictors with different horizons
p1 = observer_predictor(Gh)
p2 = observer_predictor(Gh, h=2)
p10 = observer_predictor(Gh, h=10)
p100 = observer_predictor(Gh, h=100)
bodeplot([p1, p2, p10, p100], plotphase=false, lab=["1" "" "2" "" "10" "" "100" ""])
bodeplot!(sys, ticks=:default, plotphase=false, l=(:black, :dash), lab=["sim" ""], title=["From u" "From y"])The prediction error as a function of prediction horizon approaches the simulation error.
using Statistics
hs = [1:40; 45:5:80]
perrs = map(hs) do h
yh = predict(Gh, d; h)
ControlSystemIdentification.rms(d.y - yh) |> mean
end
serr = ControlSystemIdentification.rms(d.y - simulate(Gh, d)) |> mean
plot(hs, perrs, lab="Prediction errors", xlabel="Horizon", ylabel="RMS error")
hline!([serr], lab="Simulation error", l=:dash, legend=:bottomright, ylims=(0, Inf))Validation API
predplotsimplotcoherenceplotautocorplotcrosscorplotmodelfitControlSystemIdentification.rmsControlSystemIdentification.mseControlSystemIdentification.sse
StatsAPI.predict — Functionpredict(ARX::TransferFunction, d::InputOutputData)One step ahead prediction for an ARX process. The length of the returned prediction is length(d) - max(na, nb)
Example:
julia> predict(tf(1, [1, -1], 1), iddata(1:10, 1:10))
9-element Vector{Int64}:
2
4
6
8
10
12
14
16
18predict(sys, d::AbstractIdData, args...)
predict(sys, y, u, x0 = nothing)See also predplot
yh = predict(ar::TransferFunction, y)Predict AR model
LowLevelParticleFilters.simulate — FunctionControlSystemIdentification.sse — Functionsse(x)Sum of squares of x.
ControlSystemIdentification.mse — Functionmse(x)Mean square of x.
ControlSystemIdentification.rms — Functionrms(x)Root mean square of x.
ControlSystemIdentification.fpe — Functionfpe(e, d::Int)Akaike's Final Prediction Error (FPE) criterion for model order selection.
e is the prediction errors and d is the number of parameters estimated.
ControlSystemIdentification.aic — Functionaic(e::AbstractVector, d)Akaike's Information Criterion (AIC) for model order selection.
e is the prediction errors and d is the number of parameters estimated.
See also fpe.
Video tutorials
Relevant video tutorials are available here: